- 2D experiment in 96-well plate
- cell line: PANC-1 7500 cells/well; P49; viability 81 %
- nanoparticles: GGAG:Ce@SiO2, GGAG:Ce@SiO2-RB
(new batch with RB, less pink colour)
- concentration: 0.0; 0.05; 0.5; 5.0; 50; 500; 1000 µg/mL
- other conditions:
- total killing (TK) with 10% DMSO (6 wells)
- blank MTS solution (6 wells)
- covering the plate with aluminum foil all the time; inclusive of
incubation
- growing time approx 24h
- incubation time with NPs 24h
- MTS incubation 4h
- R1 first replication
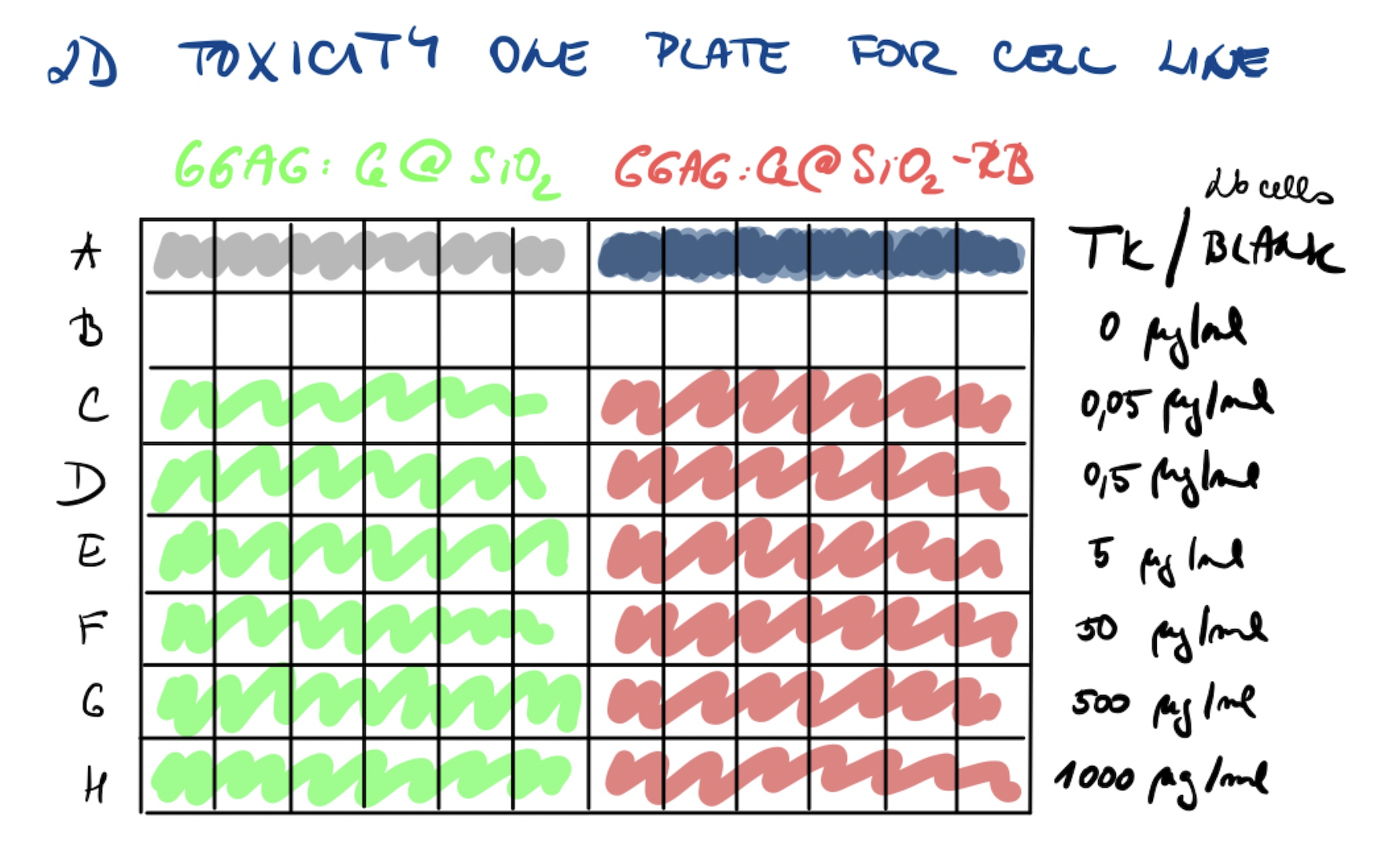 Plate map
Plate map
Experiment procedure
- 96-well plate, seeding 7500 cells/well in 100 µL culture medium
- Growing from: 19.12. 9:00; about 24h
- Removing medium
- Absorbance measurement at 485 nm (cells without any nano or
medium)
- Adding 100 µL nanoparticle’s suspension (suspension prepared before
the adding),
nanoparticles mixed by vortex before pipetting into the vials
- Covering with aluminum foil and putting to incubator
- Incubation from: 19.12. 9:45; about 24h
- Rinsing from: 20.12. 9:55
- Rinsing cells with PBS (takes about 0.5h till starting incubation
MTS)
- Absorbance measurement at 485 nm
- Adding 80 µL of medium + 20 µL CellTiter Aqueous One Solution Cell
Proliferation Assay
- 2 hours incubation
- Absorbance measurement at 485 nm
- Putting 80 µL of the viability solution to new 96-well plate for
measurement without cells
- Removing the rest of viability solution from the previous plate
- Absorbance measurement at 485 nm both plates
Data processing
- count average of the blank wells (MTS solution without cells)
- BLANK CORRECTED: subtract the average of the blank from each
well
- count average of control wells (cells without nano) = 100 %
viability
- VIABILITY %: count % viability for each well
- count average % viability for each condition
- count standard deviation of the sample (function STDEV.S)
Data
Measurement without
nanoparticles
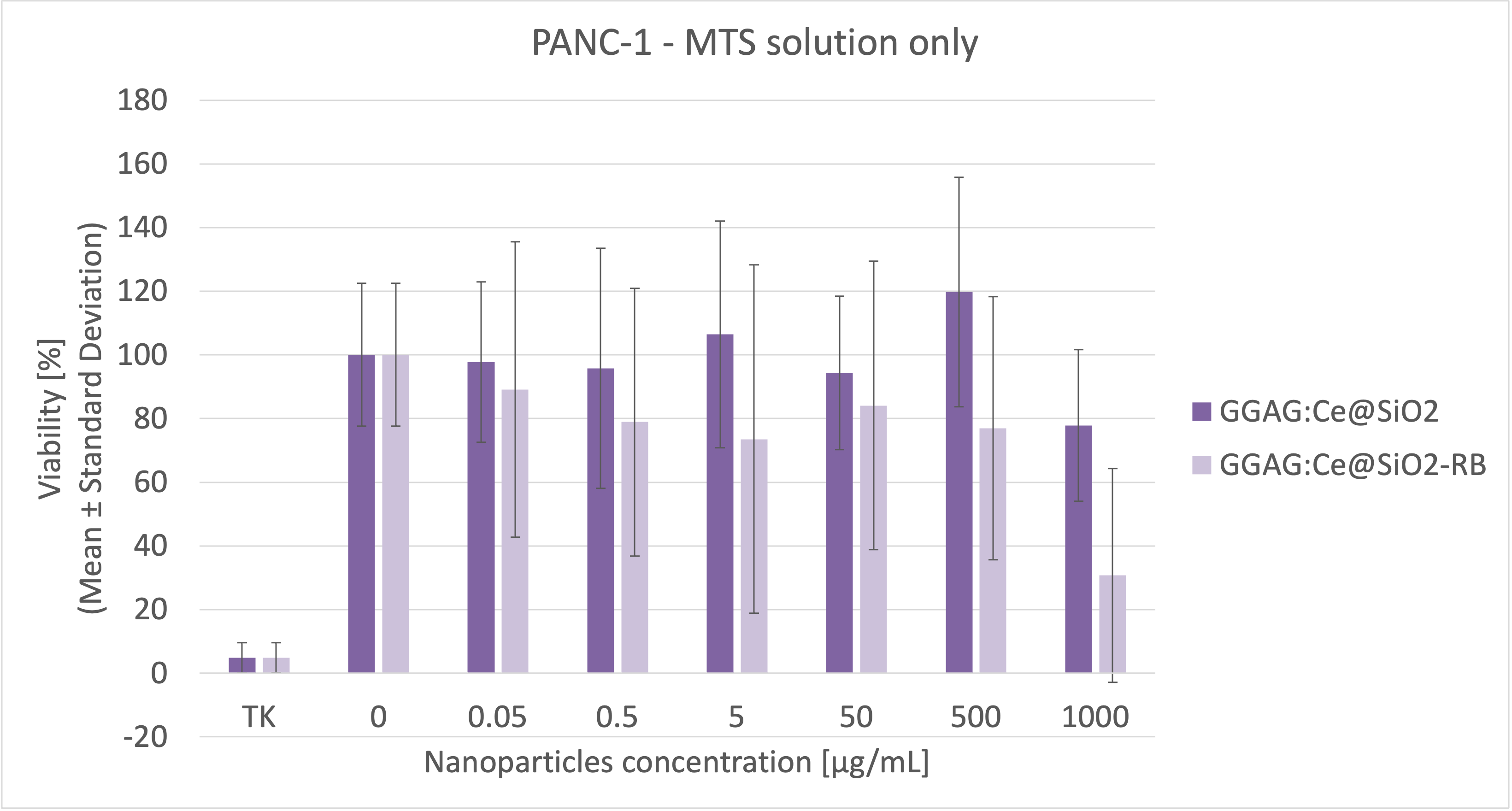 MTS
solution; 4h MTS incubation
MTS
solution; 4h MTS incubation
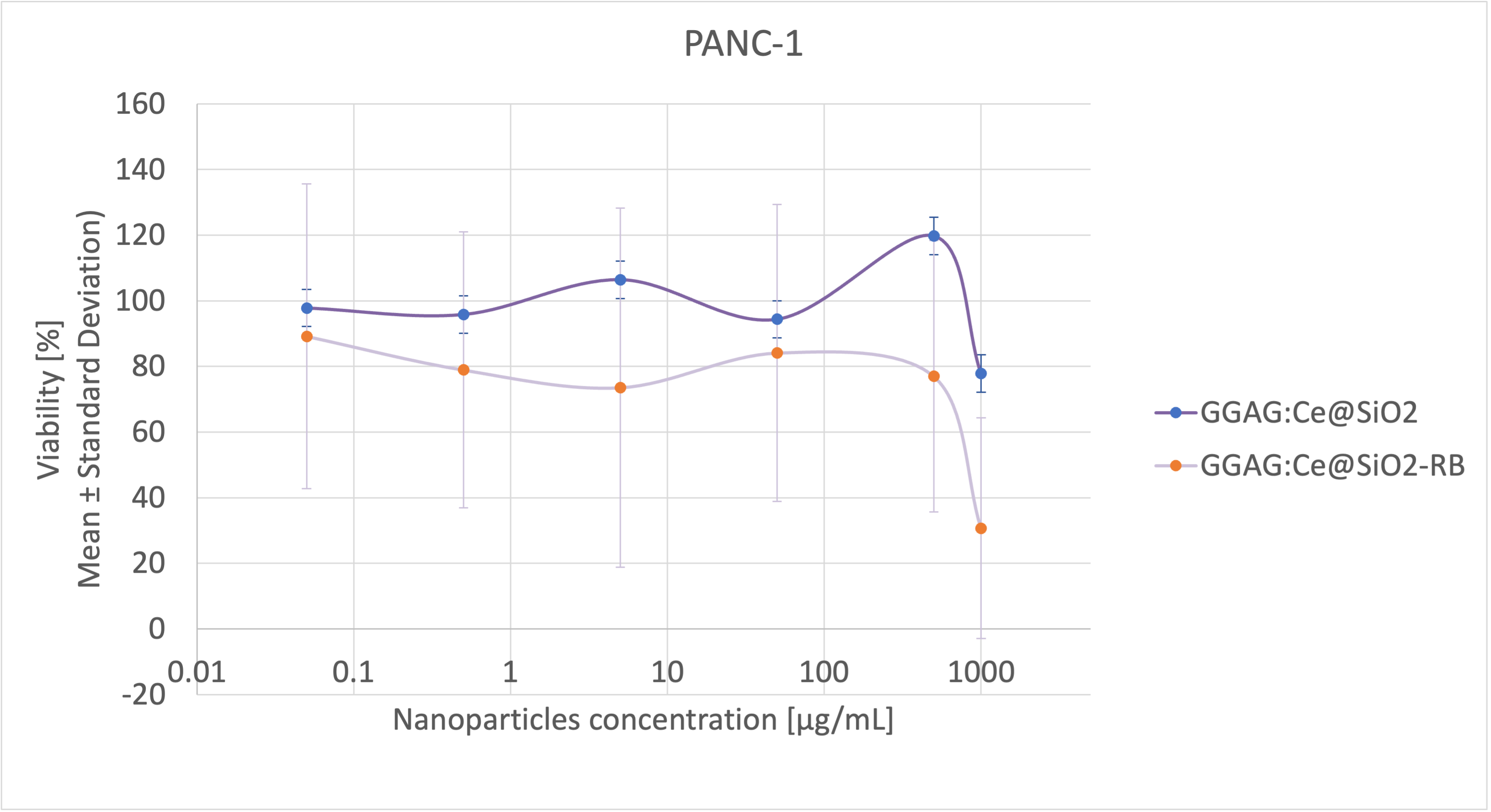 MTS
solution; 4h MTS incubation
MTS
solution; 4h MTS incubation
Viability table for all
wells
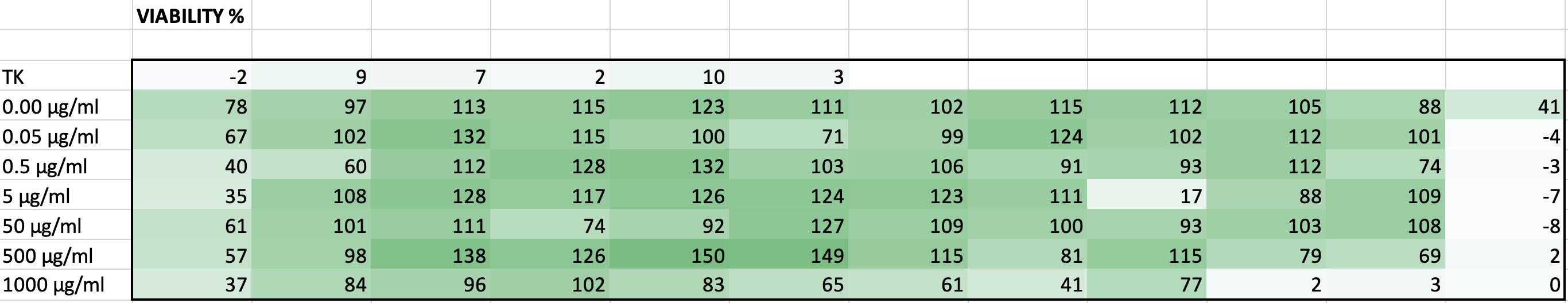 MTS solution
without cells and NPs
MTS solution
without cells and NPs




